Miscellaneous¶
Landmarking of the internal carotid artery¶
The internal carotid artery (ICA) can be classified, or landmarked, into segments. For an objective and reproducible manipulation of the ICA, we have implemented two previously published methods for landmarking: Piccinelli et al. (2011) [1], and Bogunović et al. (2012) [2]. The algorithms can also be applied to arbitrary tubular structures for automated subdivision into bends.
Although the two algorithms are well described in both articles, there is lacking information on the input parameters used to estimate curvature and torsion, which both algorithms rely on. In Kjeldsberg 2018 [3] there is a a thorough comparison between the landmarking algorithms, input parameters, and centerline smoothing methods which can help you choose the correct options for your application.
The script automated_landmarking.py includes the two following options for
automated landmarking, selected by the --algorithm flag.
- Algorithm by Piccinelli et al. (
piccinelli)- Algorithm by Bogunović et al. (
bogunovic)
Furthermore, the script includes three methods for computing
the geometric properties (curvature and torsion) of the centerline, set with
--approximation-method.
- B-Splines (
spine)- Discrete derivatives (
disc)- VMTK (
vmtk)
To perform landmarking, we will be using the model with ID C0001 from the Aneurisk database. For the commands below we assume that there is a file ./C0001/surface/model.vtp, relative to where you execute the command. To landmark the surface model, run the following command:
python automated_landmarking.py --ifile C0001/surface/model.vtp --algorithm bogunovic --approximation-method spline --nknots 8
The command will output a file C0001/surface/landmark_[ALGORITHM]_[APPROXIMATION_METHOD].particles
which contains points defining the interfaces between the segments of the vessel.
To visualize the resulting interfaces, the user may supply the argument --viz True.
In Figure 1 we show the resulting landmarks for both algorithms for case C0001.
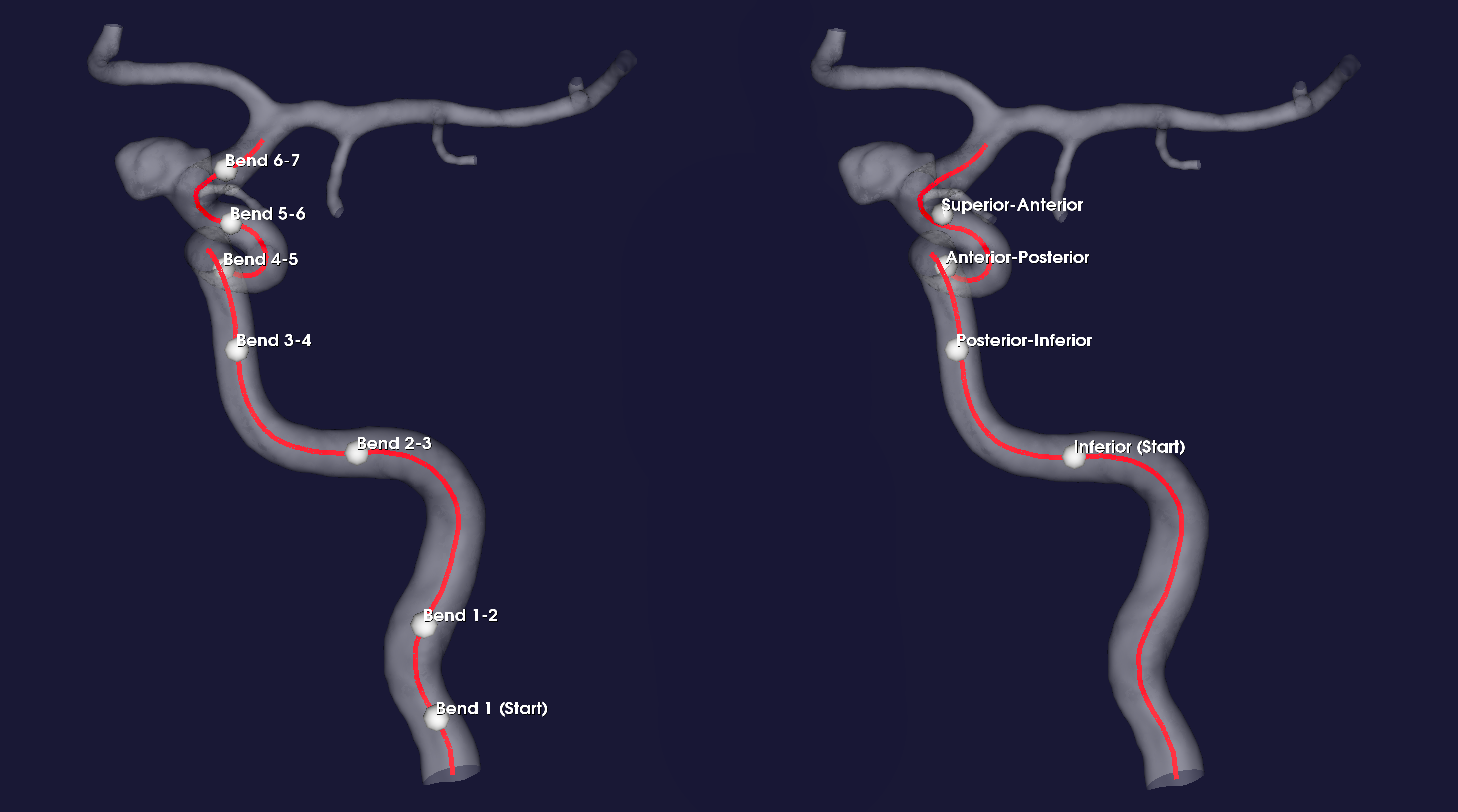
Figure 1: Landmarked geometry, with interfaces shown as white spheres along the red centerline. To the left using piccinelli, to the right using bogunovic.
Compute alpha and beta¶
Please see Tutorial: Manipulate bend for a definition of \(\alpha\) and \(\beta\).
Instead of directly setting the extent the model should be moved (--alpha and --beta),
it is more convenient to control a morphological parameter like maximum curvature, or the
angle in the bend.
The idea behind estimate_alpha_beta_values.py is to use the centerline as a
proxy for the new geometry, used to compute quantities such as curvature or bend angle.
For estimation of \(\alpha\) and \(\beta\), the script
manipulates only the centerline for a range of --alpha and
--beta values. The resulting 2D data can be fitted to a surface through cubic spline interpolation, from
which one can easily collect appropriate values for --alpha and --beta.
To estimate \(\alpha\) and \(\beta\), we will be using the model with ID C0005 from the Aneurisk database. For the commands below we assume that there is a file ./C0005/surface/model.vtp, relative to where you execute the command.
Imagine we are interested in changing the bend angle by \(\pm 10^{\circ}\). To find appropriate values for \(\alpha\) and \(\beta\), we can run the script through the following command:
python estimate_alpha_and_beta.py --ifile C0005/surface/model.vtp --quantity angle --value-change 10 --grid-size 25 --region-of-interest commandline --region-points 49.9 41.3 37.3 48 50.3 38.2
The command will output a file ./C0005/surface/model_alphabeta_values.txt
which contains two \((\alpha, \beta)\) - pairs, corresponding to the appropriate values resulting in plus and minus
the desired change in angle.
Thus we can proceed manipulation of the bend as described in Tutorial: Manipulate bend.
The algorithm can also be tweaked to get the initial value for the chosen quantity, as well as adding additional
geometric quantities to compute for.
For more information on the input parameters in the script, see automated_landmarking.automated_landmarking().
For a more detailed description of the method, please see [3].
Common¶
In the folder common we have collected utility scripts containing generic functions used by multiple scripts.
Many of the functions wrap existing vtk and vmtk functions in a more pythonic syntax,
collected in vtk_wrapper.py and vmtk_wrapper.py, respectively.
Instead of writing 6-7 lines of code to initiate a vtk-object, and set each parameter,
and the input surface, one may call one function with multiple arguments instead,
see for instance vtk_wrapper.vtk_compute_threshold().
In addition to wrapping vtk and vmtk functionality, there is also new methods for manipulating centerlines, surface models and Voronoi diagrams, collected in their respective scripts.
| [1] | Piccinelli, M., Bacigaluppi, S., Boccardi, E., Ene-Iordache, B., Remuzzi, A., Veneziani, A. and Antiga, L., 2011. Geometry of the internal carotid artery and recurrent patterns in location, orientation, and rupture status of lateral aneurysms: an image-based computational study. Neurosurgery, 68(5), pp.1270-1285. |
| [2] | Bogunović, H., Pozo, J.M., Cárdenes, R., Villa-Uriol, M.C., Blanc, R., Piotin, M. and Frangi, A.F., 2012. Automated landmarking and geometric characterization of the carotid siphon. Medical image analysis, 16(4), pp.889-903. |
| [3] | (1, 2) Kjeldsberg, Henrik Aasen. Investigating the Interaction Between Morphology of the Anterior Bend and Aneurysm Initiation. MS thesis. 2018. |